Research
Research Interests
Structural biology, cryo-EM, sample preparation, automation, molecular complexes, big data
Publications and preprints
Research Experience
Scientist, Simons Electron Microscopy Center, New York Stuctural Biology Center, 2020-present
Vitrocam
 With a focus on improving automation and efficiency in cryo-EM sample preparation, we built the Vitrocam, a simple and cost-effective Raspberry Pi camera, to image grids in the Vitrobot during plunging. By providing information on grid and ice quality during plunge freezing with the Vitrocam, the sample preparation feedback loop can be shortened, compared to otherwise costly and time-consuming screening in an electron microscope.
With a focus on improving automation and efficiency in cryo-EM sample preparation, we built the Vitrocam, a simple and cost-effective Raspberry Pi camera, to image grids in the Vitrobot during plunging. By providing information on grid and ice quality during plunge freezing with the Vitrocam, the sample preparation feedback loop can be shortened, compared to otherwise costly and time-consuming screening in an electron microscope.
Ice thickness
We measured the effect of ice thickness and a range of microscope configurations on resolution in single particle analysis.
Access and Training
I pioneered the grid preparation and screening program at our NIH-funded National Center for Cryo-EM Access and Training (NCCAT). Through this program, users with limited cryo-EM access or expertise can gain access and training to prepare their samples for high resolution cryo-EM imaging.
Skills
Single particle cryo-EM training and consultation, microscope operations, sample preparation and optimization for cryo-EM, Python and Jupyter lab
Research Associate, Wigley Lab, Imperial College London, 2018-2020
Biological system
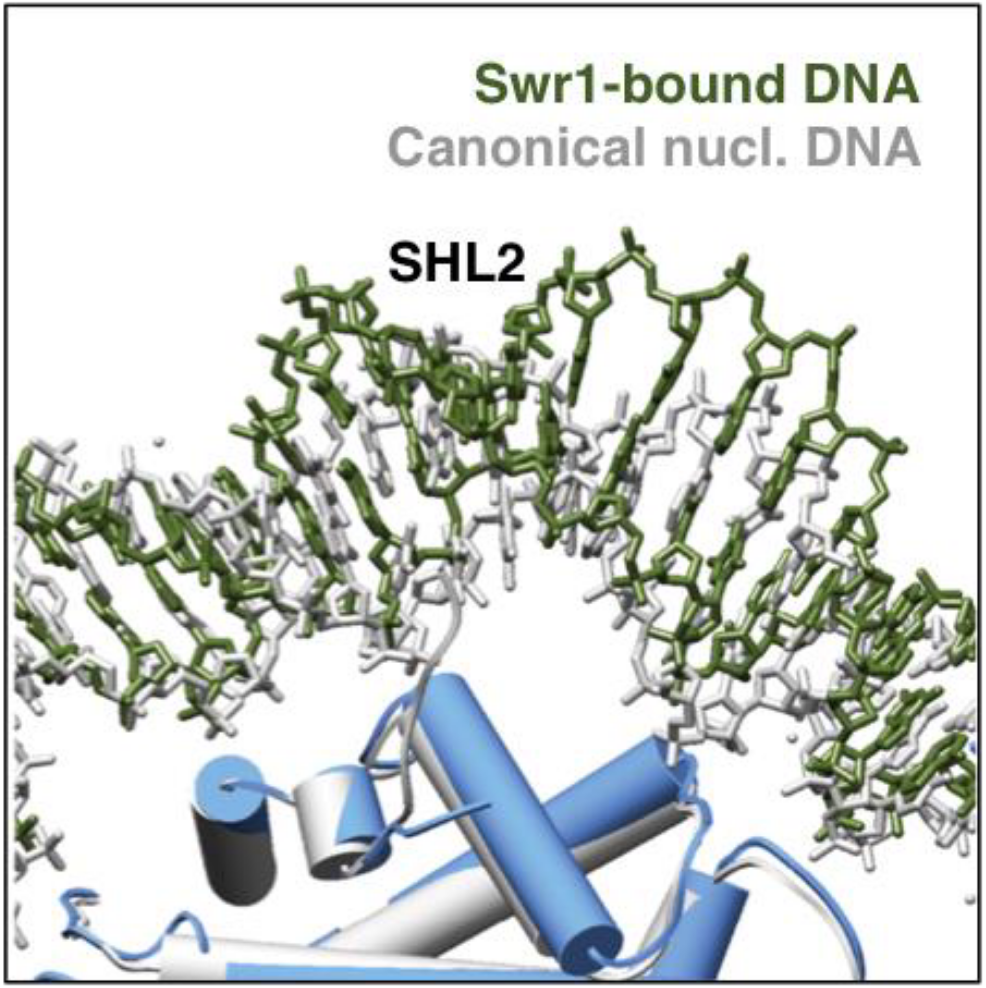 We studied SWR1, a yeast chromatin remodeling complex involved in histone exchange. Through ATP catalysis, SWR1 removes the canonical H2A-H2B histone dimer from the nucleosome and replaces it with the Htz-H2B variant dimer.
We studied SWR1, a yeast chromatin remodeling complex involved in histone exchange. Through ATP catalysis, SWR1 removes the canonical H2A-H2B histone dimer from the nucleosome and replaces it with the Htz-H2B variant dimer.
Impact
We determined the first high resolution structure of SWR1 by cryo-EM. This revealed the molecular architecture of SWR1 and the first steps of SWR1 binding to the nucleosome. In collaboration with single molecule biophysicists, we described the dynamics in nucleosomal DNA during SWR1 binding and histone exchange.
Tools
Single particle cryo-EM, fluorescence-based enzyme kinetics, microscale thermophoresis, electromobility shift assay, insect cell culture and Multibac cloning, protein expression and purification
Research Fellow, Sandin lab, Nanyang Technological University, Singapore, 2015-2017
Impact
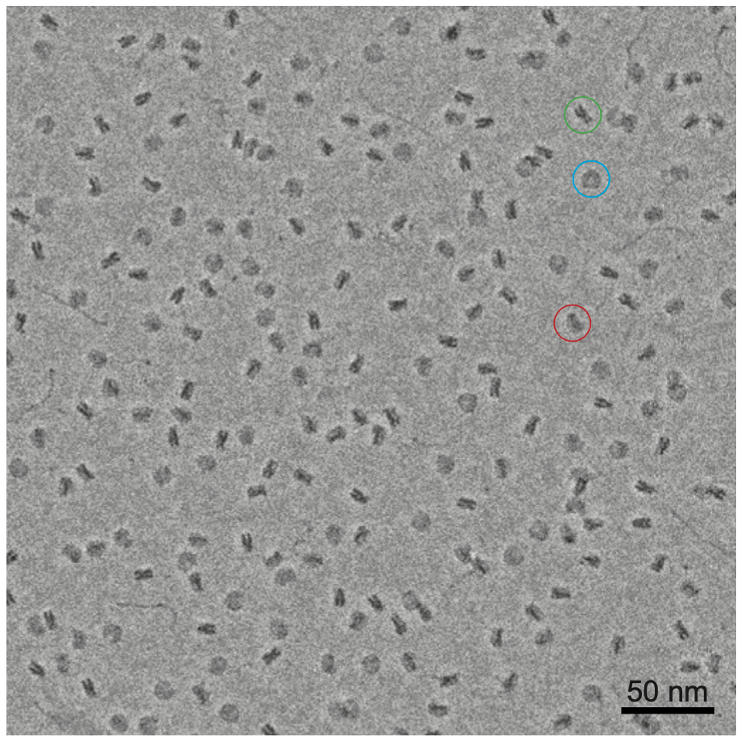 We applied the Volta phase plate to single particle cryo-EM to show that imaging DNA and reconstructing small 200 kDa nucleosome particles was possible.
We applied the Volta phase plate to single particle cryo-EM to show that imaging DNA and reconstructing small 200 kDa nucleosome particles was possible.
Tools
Single particle cryo-EM, Vola phase plate, mammalian cell culture
Doctoral Research, Davey lab, Nanyang Technological University, Singapore, 2010-2014
Biological system
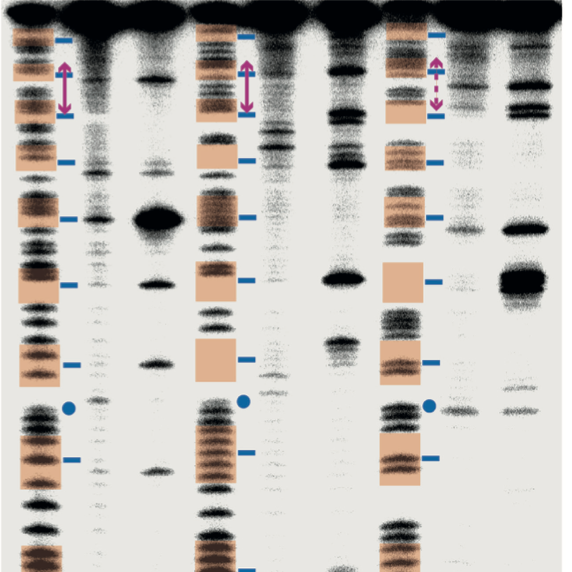 We studied the structures and functions of nucleosomes, the basic repeating unit of chromatin. Nucleosomes are responsible for compacting, organizing, and providing access to DNA in eukaryotic cells.
We studied the structures and functions of nucleosomes, the basic repeating unit of chromatin. Nucleosomes are responsible for compacting, organizing, and providing access to DNA in eukaryotic cells.
Impact
We designed and characterized novel platinum-based agents that target unique DNA structural motifs in the nucleosome, furthering our ability to design cancer-specific DNA-binding molecules. We also provided insight into the mechanisms underpinning DNA binding to the histone octamer by determining X-ray crystal structures of nucleosomes reconstituted with a variety of DNA sequences.
Skills
X-ray crystallography, DNA footprinting, tyrosine fluorescence quenching, surface plasmon resonance, bacterial cell culture and cloning
Education
2010-2014 PhD, Biological Sciences, Nanyang Technological University, Singapore
2006-2010 BSc (Hons), Biological Sciences, Nanyang Technological University, Singapore